In our group we have activities within the fields of photosynthesis, synthetic biology and biophysics.
Within photosynthesis:
- Role of pili in the acquisition of metals from metal oxides by cyanobacteria
- Photosynthetic machinery of heterokont algae
Within synthetic biology:
Within biophysics:
Photosynthesis
Photosynthesis is arguably the most important biological process on Earth. This process is responsible for the oxygen in our atmosphere, and the organic molecules produced by photosynthetic organisms fuel animal life. We encounter a diversity of photosynthetic organisms, which have photosynthetic machineries that have evolved to perform in a large variety of environments, both in the oceans and on land.
One focus of PhotoSynLab is to find out how the photosynthetic machinery is adapted to different environments. We investigate cyanobacteria, green algae, and heterokont algae, including diatoms. In order to obtain comprehensive insights into the photosynthetic machinery, we utilize molecular biology and bioinformatics, in combination with biochemical and biophysical techniques.
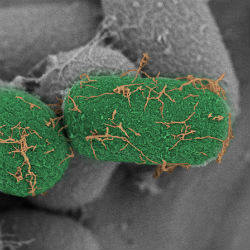

Iron and manganese are two metals that are crucial for photosynthetic machinery in cyanobacteria. However, these metals are complexed into metal oxides in most environments, which makes them insoluble and thus unavailable for cyanobacteria. How do cyanobacteria overcome metal limitation? There is mounting evidence that bacterial pili are able to couple metabolically-generated electrons to the extracellular environment. Electron donation via “nanowires” to metal oxides renders metals bio-available. We characterizing the role of pili in metal acquisition in the cyanobacteria Synechococcus sp. PCC 7002 and Synechococystis sp. PCC 6803.
Photosynthetic machinery of heterokont algae
Heterokont algae comprise a diverse group of organisms including Bacillariophyceae (diatoms) and Eustigmatophyceae. The photoprotective mechanisms that these organisms employ include energy dissipation that is mediated by the conversion between two different forms of carotenoids. In addition, dynamic redistribution of light energy between the two types of photosystems seems to play a role in avoiding photodamage. Differentiating between carotenoid conversion and dynamic light distribution is a current focus of the laboratory.
Synthetic biology
Synthetic biology is a multidisciplinary field that merges life sciences with engineering. In our lab we are developing novel synthetic biology tools to create microbial “cell factories” that are tailored towards manufacturing of desired products. We utilise a variety of photosynthetic and non-photosynthetic bacteria and eukaryotes as host organisms such as Corynebacterium glutamicum, Escherichia coli, Pseudomonas putida KT2440, psychrophilic Pseudomonas, Psychrobacter spp., Saccharomyces cerevisiae, Streptomyces albus, S. coelicolor, S. lividans, Synechococcus sp. PCC 7002, Synechocystis sp. PCC 6802, Thermus thermophilus.
We develop platforms that allow the design of artificial promoter and 5` UTR sequences that are tailored towards the gene of interest in the host of interest. In order to achieve rapid progress we also utilize a variety of techniques, including high-throughput screening, single-cell array analysis and microfluidics.

PhotosynLab is developing and characterising molecular tools that enable the rapid construction of genetic elements into functional units, such as plasmids and DNA integration sites. In addition, we develop conjugation-based DNA delivery systems that accomplish high yield DNA integration into the genomes of eukaryotic organisms.
Artificial promoter and 5´ UTR design
Gene and protein expression is at the heart of synthetic biology. PhotoSynLab is developing expression tools that allow for the production of proteins at desired levels and under a large variety of conditions. To this end we design artificial promoters and 5′ UTRs that lead to the desired level of gene and protein expression in a wide range of microorganisms including Corynebacterium glutamicum, Escherichia coli, Pseudomonas putida KT2440, Saccharomyces cerevisiae, Streptomyces albus, S. lividans, Thermus thermophilus.
Collaborations
Within the synthetic biology we do collaborate with:
- Anil Wipat – Director of SBOL, School of Computing, Newcaste, England
- Christian Rückert, Jörn Kalinowski – Technology Platform Genomics, Center for Biotechnology, Bielefeld University, Germany
Biophysics

Alginate CLEX-microfluidics
(Image: Swapnil Bhujbal)

Mono-disperse droplet generator
(Image: Swapnil Bhujbal)
Microfluidics
Microfluidics in simple words is controlling or manipulating the fluids at the micrometer scale, which has offered a great number of opportunities in life science research. Microfluidics channels are produced with soft lithography techniques. Our lab uses microfluidics platform combined with alginate competitive ligand exchange crosslinking method to extract novel microbial genomes while preserving single-cell resolution, with increased throughput and sensitivity while at the same time reducing the cost.
Details
Briefly the protocol involves spin-coating of silicon wafers with UV sensitive photoresist, UV exposure through mask or maskless aligner with desired microfluidics design files, development of uncured resist to form a mold, polydimethylsiloxane(PDMS) elastomer casting, baking, punching and bonding. Our lab uses microfluidics platform combined with alginate Competitive Ligand EXchange crosslinking (CLEX) method to extract novel microbial genomes while preserving single-cell resolution, with increased throughput and sensitivity while at the same time reducing the cost.
Collaborations
Within the microfluidics we do collaborate with:
- Bjørn Torger Stokke – Department of Physics, NTNU, Norway
- Liisa van Vliet, Florian Hollfelder – Department of Biochemistry, University of Cambridge, England

Poly-L-lysine fluorescein isothiocyanate (PLL-FITC) single-cell arrays
(Image: Swapnil Bhujbal)

PLL-FITC staining of alginate beads
(Image: Swapnil Bhujbal)
Single-cell arrays
Bacterial heterogeneity or cell-to-cell variability cannot be answered by traditional microbiological experimental approaches which generally reveal population-wide statistics. Our lab uses soft lithography based microcontact-printing techniques to answer cellular heterogeneity.
Details
Briefly first we generate PDMS stamps for patterning, then we simultaneously coat a glass slide with a polymer which resist bacterial adhesion and PDMS stamps which promote bacterial adhesion, before the excess is dried off, the PDMS stamp is placed patterned side down on the polymer coated glass surface, giving rise to chemically patterned surface. We then subsequently incubate the bacteria of interest in solution on to a patterned surface followed by rinsing off non-adhered bacteria, which therefore results in bacteria adhesion only to selected areas of glass substrate thus generating a single-cell array. We also combine microcontact-printing technique to adhere alginate beads. The immobilized beads combined with micromanipulator serves as platform for metagenomics screenings.
Collaborations
Within the single-cell arrays we do collaborate with:
- Marit Sletmoen – Department of Biotechnology, NTNU, Norway
Below is an overview of our ongoing research activities.
RCN – Research Council of Norway
EPSRC – The Engineering and Physical Sciences Research Council, UK
EU – European Union
NTNU – Norwegian University of Science and Technology
MT – Master’s Thesis
 Seed Funding – NTNU
Seed Funding – NTNU
 NanoWires – RCN
NanoWires – RCN BioSmart – RCN
BioSmart – RCN MetaFluidics – EU-H2020
MetaFluidics – EU-H2020 Robotic Infrastructure – NTNU
Robotic Infrastructure – NTNU SUPERAPP – NTNU-Discovery (Pilot)
SUPERAPP – NTNU-Discovery (Pilot) Molecular Foundry User Grant
Molecular Foundry User Grant EPSRC – UK – Design the Future 2: Thinking Soils
EPSRC – UK – Design the Future 2: Thinking Soils